New day, new genetics analysis work flow. This time I’m going to use GenePop, a standard program that (apparently) does everything I need it to do!
Checking 2016/2017 Fidalgo Bay raw data for correct binning
Crystal rounded the raw microsat data for the Fidalgo Bay 2016-hatchery and 2017-wild data. She provided both raw and rounded data. Before moving forward with the rounded data, I’ll check out the binning method she used.
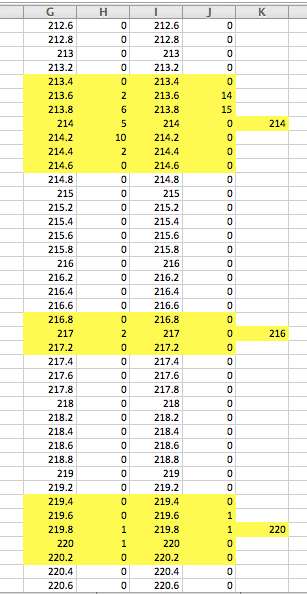
In the Excel file Olympic Oyster NFH_NFW (1).xlsx she includes data from both wild and hatchery NF samples. She houses raw data for each locus in separate tabs, creates a list of “bins” at 0.2 increments, calculates frequencies for each bin and visualizes with histograms.

Then, using the frequency distributions she assigned alleles, for example:
 |
 |
One question I have is regarding the assignment of all even-numbered alleles for Oly10, Oly11 & Oly12, while alleles are odd for Oly13, Oly15 & Oly19.
I also noticed that Oly18 data was initially processed, then not completed nor included in the “rounded” tab. I emailed Crystal to see what’s up (I presume it was an oversight).
Next step is to export the data into a GenePop format. GenePop is one of the most commonly used programs used to analyze microsatellite data. There are several ways to use GenePop: on the web, at the command line, and in R. I like to work in R. I could not find an R-based function to convert .csv format to GenePop format, however thre is an Excel plug-in caled GenAlEx that one can use. I download version 6.503 (Dec 5, 2016). Then, I merged the wild and hatchery data into one spreadsheet. I also found online that the commonly used “genind” format has a few key formatting requirements, which I point out in the following screenshot:

With this merged file open, I also opened the GenAlEx program. Then, I used the GenAlEx plug-in to export the file as a GenPop formatted .txt file:

A window pops up, which should automatically ID the #loci & #samples if you formatted the spreadsheet like I did; I edited the “Title” to include 2016/2017 info.

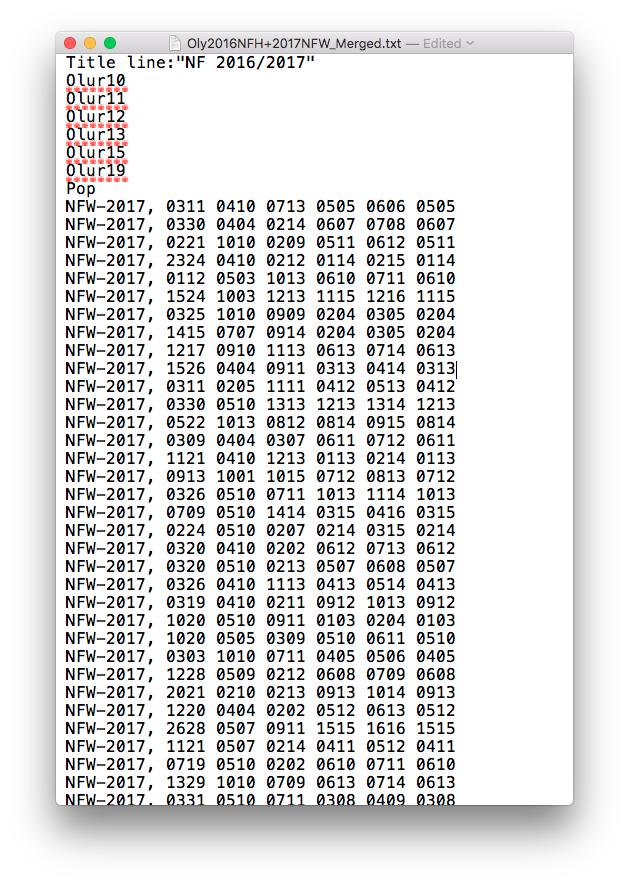
Saved the file as a .txt file under Oly2016NFH+2017NFW_Merged.txt; here’s what the resulting PopGen formatted file looks like:
from LabNotebook http://ift.tt/2DywTRd
via IFTTT
