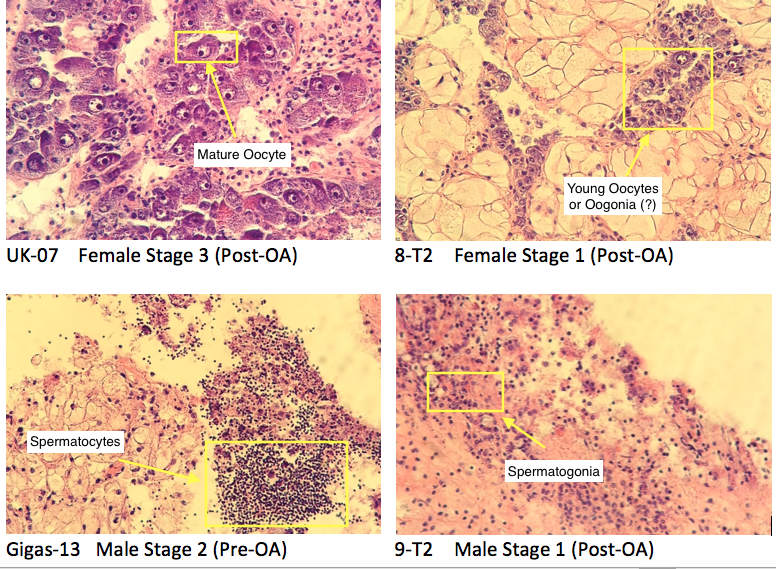
(1) I figured out a better way to image the larvae. I dump the ethanol in the tube into a 80ml beaker. Then, I dump it back into the tube. This dislodges all of the stuff at the bottom. Then I dump the ethanol back in the beaker, and it ends up containing the larvae, leaving behind the larger bits of junk. I wait for the larvae to settle, then pipette off the ethanol until I get to the larvae.
(2) Started the Analysis of the 2015 Oyster seed data. Emma performed the Pecan step, so I started in at Step 4, using the .blib file that she created. I am stuck at Step 4e . We need an isolation scheme file from Step 3b:

We asked Emma to help us out. And here is what she provided: GitHub Issue
(3) Titrator: I worked through the SOP to calibrate the pH probe. It seemed to work well. Then Steven and I tried out the CSUN method from Hollie on pH buffers – just to try out the method. It seems a little odd to me because it did each sample one at a time. I thought that it would run like that pH calibration method where it went from sample to sample automatically.