I am glad to be part of this group.
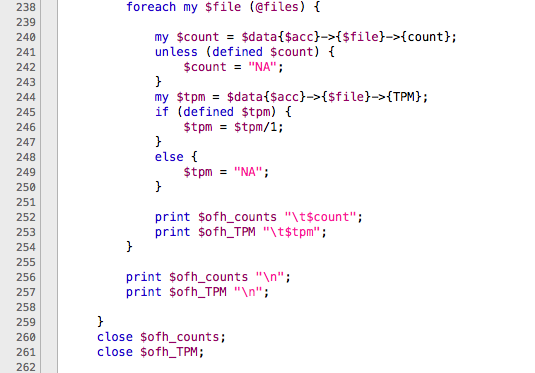
Well, Working on trinity (trinityrna-2.2.0) specifically in abundance estimation of sequences using RSEM package (Before to run differential expression analyses). I had two files called RSEM.isoforms.results and RSEM.genes.results. Both of them are matrices with values of length, effective length of genes, expected count, TPM (transcripts per million) and FPKM (fragments per kilo base million). The TPM and FPKM values are used by the script abundance_estimate_to_matrix.pl to estimate the matrices for differential expression, but, there is a problem generating them using the RSEM.genes.results. The part of the script
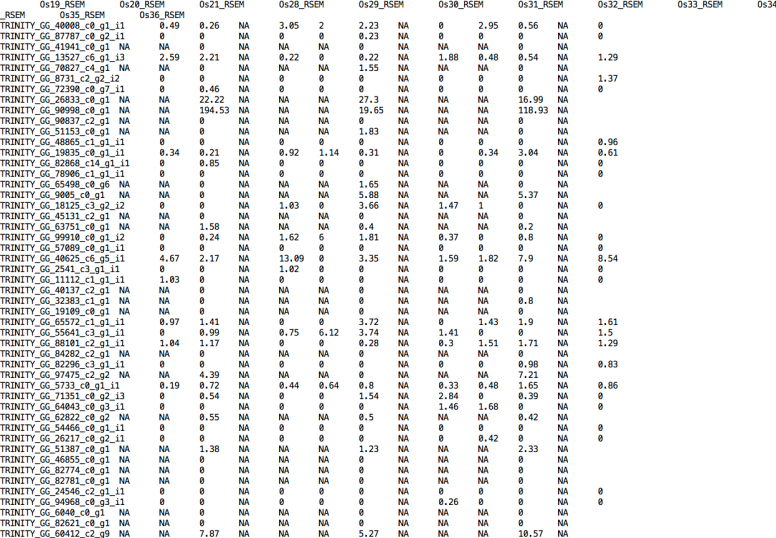
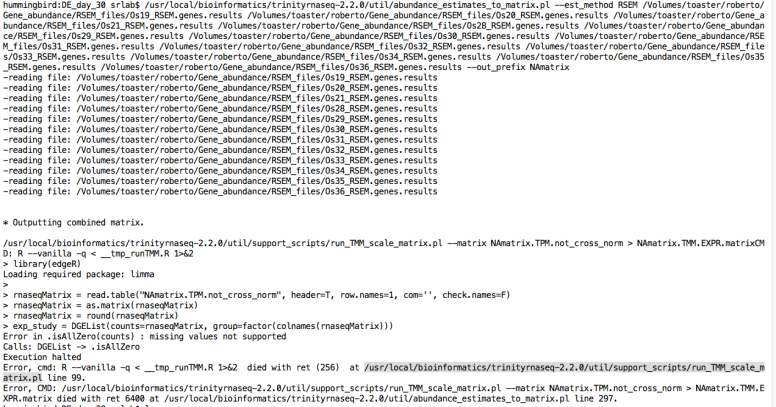
At line 242 and 249, writes “NA” for absent gene IDs and this makes an error to create a matrix used for differential expression analyses because the script just recognize numeric values.
But rewriting the script changing NA by 0 helps to create the matrices but differential expression analyses had different results than using RSEM.isoforms.results.
I can’t see the sense for this part. I mean why should be NA instead of 0? even if it needs numeric values and does this affects the differences using genes.results (where I found 10 differentially expressed genes) and isoforms.results (where I found 384 differentially expressed transcripts)? Now I am trying to have the annotation for those 10 genes and compare their ontology with the isoforms annotation.